Case Study 1: Simulation Recovery
David Hodgson
2025-12-03
cs1_sim_recovery.RmdR Code Description
This document describes the steps and components of the provided R code for creating and running serological models using simulated data. The code defines observational models, antibody kinetics models, and runs inference using MCMC methods.
Step 2: Read Simulated Data
This data has been generated using serosim. The code the
generate this data can be found at in the folder R/serosim
in the package. The data is generated using the cesCOP and
cesNoCOP models.
sim_model_cop <- readRDS(file = "cesCOP_inputs.RDS")
sim_res_cop <- readRDS(file = "cesCOP_sim_data_0.5.rds")
#sim_model_cop <- readRDS(file = system.file("extdata", "vig_data", "cesCOP", "cesCOP_inputs.RDS", package = "serojump"))
#sim_res_cop <- readRDS(file = system.file("extdata","vig_data", "cesCOP", "cesCOP_sim_data_0.5.RDS", package = "serojump"))
#modeli <- readRDS(file = "cesCOP_inputs.RDS")
#modeli <- readRDS(file = "cesCOP_sin_data_0.5.RDS")
data_titre_model <- sim_res_cop$observed_biomarker_states %>% select(i, t, value) %>% rename(id = i, time = t, titre = value)
data_titre_model <- data_titre_model %>% mutate(biomarker = "sVNT") %>% as.data.frame %>% rename(sVNT = titre)
# Check the entries are sensible
check_sero_no_single_entries(data_titre_model)## No single entries in data_sero!,
check_sero_timings(data_titre_model)## No individuals with less than 15 days in the study!Step 3: Define Likelihood and Kinetics Functions
obsLogLikelihood <- function(titre_val, titre_est, pars) {
ll <- dnorm(titre_val, titre_est, pars[1], log = TRUE)
}
infSerumKinetics <- function(titre_est, timeSince, pars) {
a <- pars[1]
b <- pars[2]
c <- pars[3]
if (timeSince < 14) {
titre_est <- titre_est + log(exp(a) + exp(c)) * (timeSince) / 14;
} else {
titre_est <- titre_est + log(exp(a) * exp(-b/10 * (timeSince - 14)) + exp(c));
}
titre_est
}
noInfSerumKinetics <- function(titre_est, timeSince, pars) {
titre_est_log <- titre_est - pars[1] * (timeSince)
titre_est_log <- max(0, titre_est_log)
titre_est_log
}Step 4: Define the Observational and Kinetics Models
# Define the biomarkers and exposure types in the model
biomarkers <- c("sVNT")
exposureTypes <- c("none", "inf")
exposureFitted <- "inf"
# Define the observational model
observationalModel <- list(
names = c("sVNT"),
model = makeModel(
addObservationalModel("sVNT", c("sigma"), obsLogLikelihood)
), # observational model,
prior = bind_rows(
addPrior("sigma", 0.0001, 4, "unif", 0.0001, 4)
)
)
# Define the antibody kinetics model
abkineticsModel <- list(
model = makeModel(
addAbkineticsModel("none", "sVNT", "none", c("wane"), noInfSerumKinetics),
addAbkineticsModel("inf", "sVNT", "inf", c("a", "b", "c"), infSerumKinetics)
),
prior = bind_rows(
addPrior("wane", 0.0, 0.01, "unif", 0.0, 0.01), # observational model
addPrior("a", -6, 6, "norm", 2, 2), # ab kinetics
addPrior("b", 0, 1, "norm", 0.3, 0.05), # ab kinetics
addPrior("c", 0, 4, "unif", 0, 4) # ab kinetics
)
)
model_cop <- createSeroJumpModel(
data_sero = data_titre_model,
data_known = NULL,
biomarkers = biomarkers,
exposureTypes = exposureTypes,
exposureFitted = exposureFitted,
observationalModel = observationalModel,
abkineticsModel = abkineticsModel)## OUTLINE OF INPUTTED MODEL
## There are 1 measured biomarkers: sVNT
## There are 2 exposure types in the study period: none, inf
## The fitted exposure type is inf
## PRIOR DISTRIBUTIONS
## Prior parameters of observationalModel are: sigma
## Prior parameters of abkineticsModel are: wane, a, b, c
## No single entries in data_sero!,
## No individuals with less than 15 days in the study!
## Exposure rate is not defined over the time period. Defaulting to uniform distribution between 1 and 119 .Step 4B: Prerun sanity plots
Before running the whole model it is good to check the data and the
priors. This can be done using a suit of functions
plotPriors function.
p1 <- plotSero(model_cop)
p2 <- plotPriorPredictive(model_cop)## Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
## ℹ Please use `linewidth` instead.
## ℹ The deprecated feature was likely used in the serojump package.
## Please report the issue to the authors.
## This warning is displayed once every 8 hours.
## Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
## generated.
p3 <- plotPriorInfection(model_cop)
p1 / p2## Ignoring unknown labels:
## • colour : "Exposure type"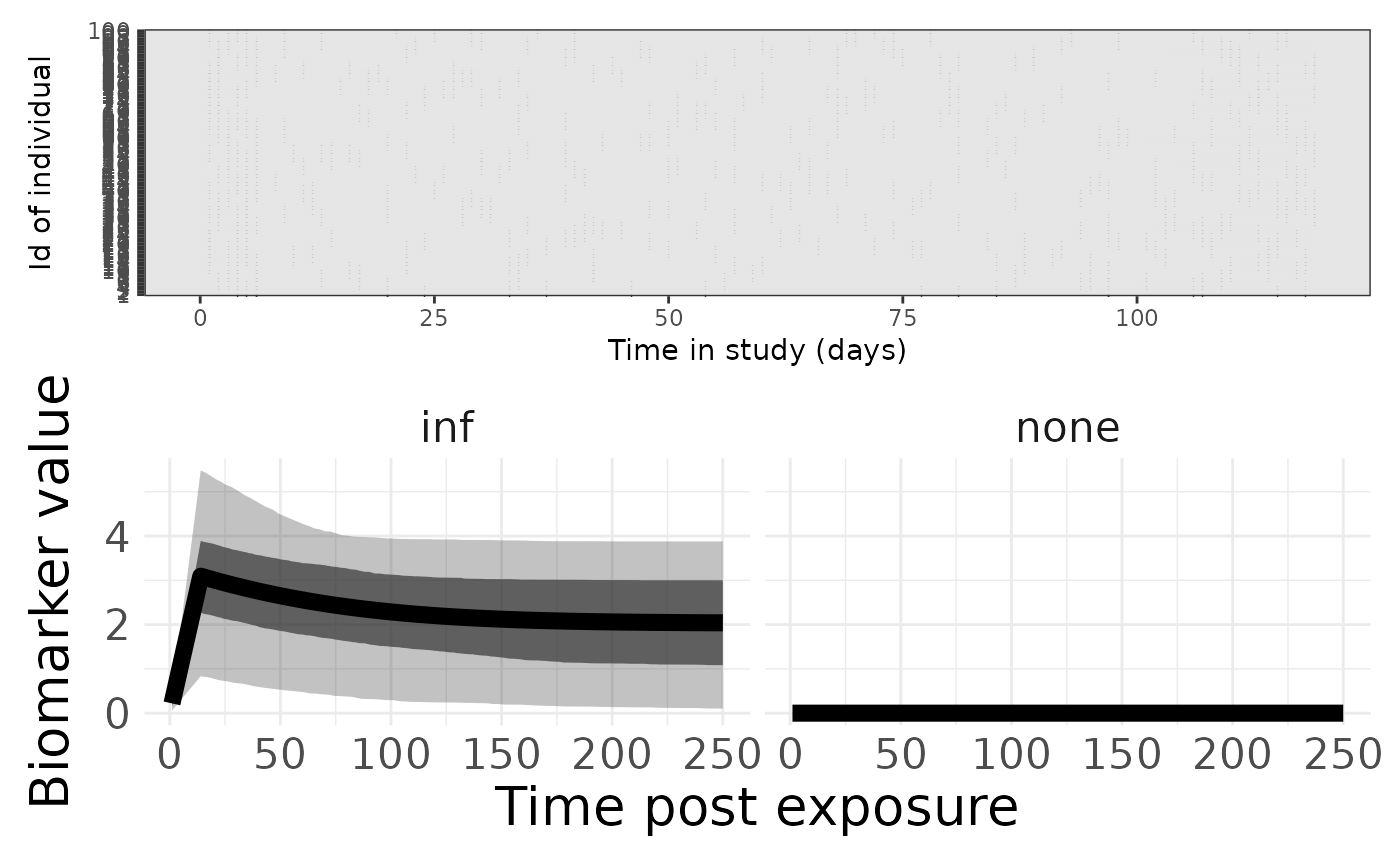
p3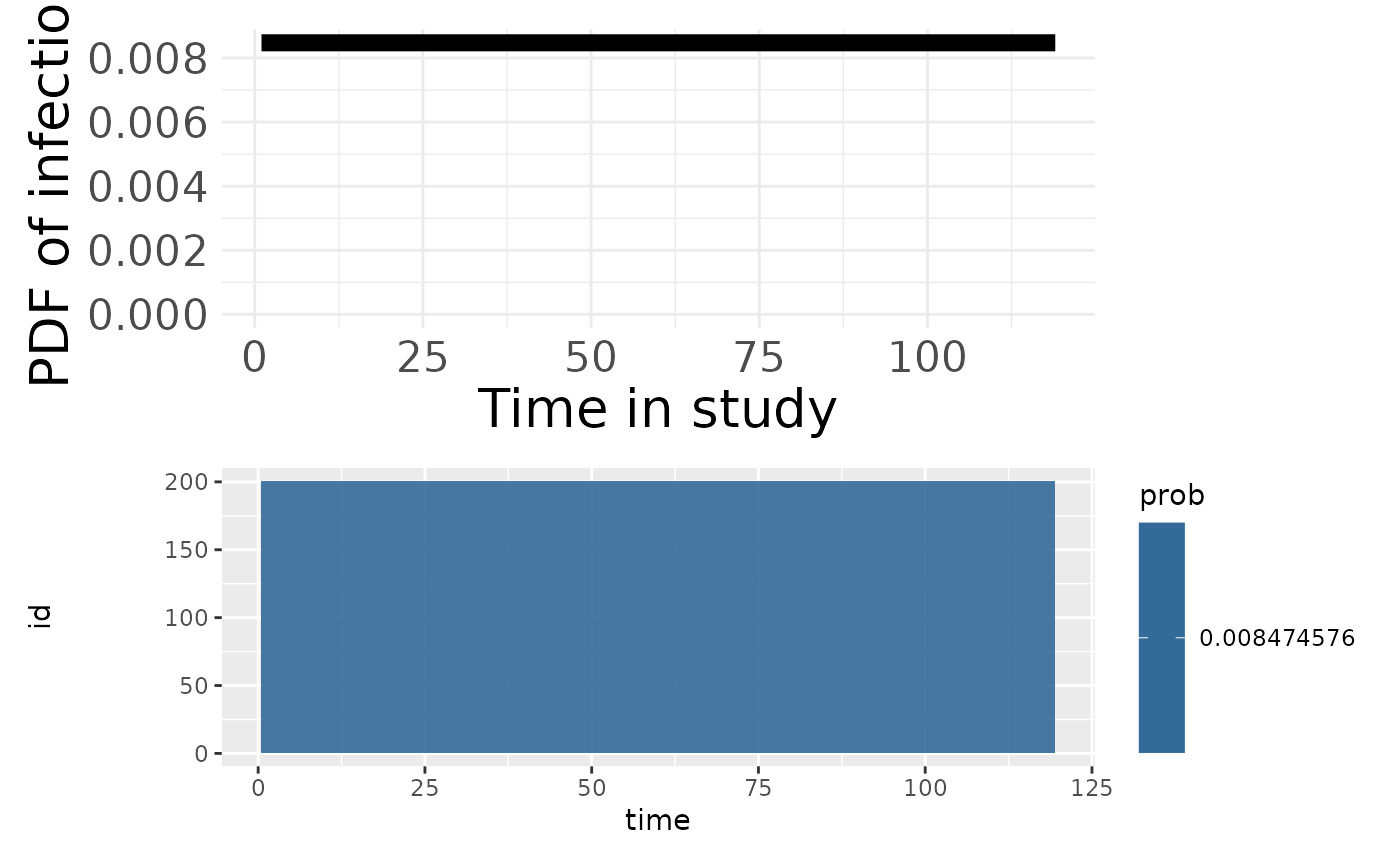
Step 5: Define the Complete Model
sim_model_no_cop <- readRDS(file = "cesNoCOP_inputs.RDS")
sim_res_no_cop <- readRDS(file = "cesNoCOP_sim_data_0.5.rds")
#modeli <- readRDS(file = system.file("extdata", "vig_data", "cesNoCOP", "cesNoCOP_inputs.RDS", package = "serojump"))
#res <- readRDS(file = system.file("extdata", "vig_data", "cesNoCOP", "cesNoCOP_sim_data_0.5.RDS", package = "serojump"))
data_titre_model <- sim_res_no_cop$observed_biomarker_states %>% select(i, t, value) %>% rename(id = i, time = t, titre = value)
data_titre_model <- data_titre_model %>% mutate(biomarker = "sVNT") %>% as.data.frame %>% rename(sVNT = titre)
model_no_cop <- createSeroJumpModel(
data_sero = data_titre_model,
data_known = NULL,
biomarkers = biomarkers,
exposureTypes = exposureTypes,
exposureFitted = exposureFitted,
observationalModel = observationalModel,
abkineticsModel = abkineticsModel)## OUTLINE OF INPUTTED MODEL
## There are 1 measured biomarkers: sVNT
## There are 2 exposure types in the study period: none, inf
## The fitted exposure type is inf
## PRIOR DISTRIBUTIONS
## Prior parameters of observationalModel are: sigma
## Prior parameters of abkineticsModel are: wane, a, b, c
## No single entries in data_sero!,
## No individuals with less than 15 days in the study!
## Exposure rate is not defined over the time period. Defaulting to uniform distribution between 1 and 119 .Step 5B: Prerun sanity plots
Before running the whole model it is good to check the data and the
priors. This can be done using a suit of functions
plotPriors function.
p1 <- plotSero(model_no_cop)
p2 <- plotPriorPredictive(model_no_cop)
p1 / p2## Ignoring unknown labels:
## • colour : "Exposure type"
rj_settings <- list(
numberChainRuns = 4,
iterations = 400000,
burninPosterior = 200000,
thin = 1000
)
save_info_cop <- list(
file_name = "simulated_data",
model_name = "cop"
)
save_info_no_cop <- list(
file_name = "simulated_data",
model_name = "no_cop"
)
# Run these but take a while
#model_summary_cop <- runSeroJump(model_cop, rj_settings, save_info = save_info_cop)
#model_summary_no_cop <- runSeroJump(model_no_cop, rj_settings, save_info = save_info_no_cop)
model_summary_cop <- readRDS("cop_model_summary.RDS")
model_summary_no_cop <- readRDS("no_cop_model_summary.RDS")
plotMCMCDiagnosis(model_summary_cop, save_info = save_info_cop)
# takes ages to run these so comment out!
#plotPostFigsSim(model_summary_cop, sim_model_cop, sim_res_cop, save_info = save_info_cop)
plotMCMCDiagnosis(model_summary_no_cop, save_info = save_info_no_cop)
# takes ages to run these so comment out!
#plotPostFigsSim(model_summary_no_cop, sim_model_no_cop, sim_res_no_cop, save_info = save_info_no_cop)