Simulate incidence curve using Gaussian Process
Source:R/exposure_models.R
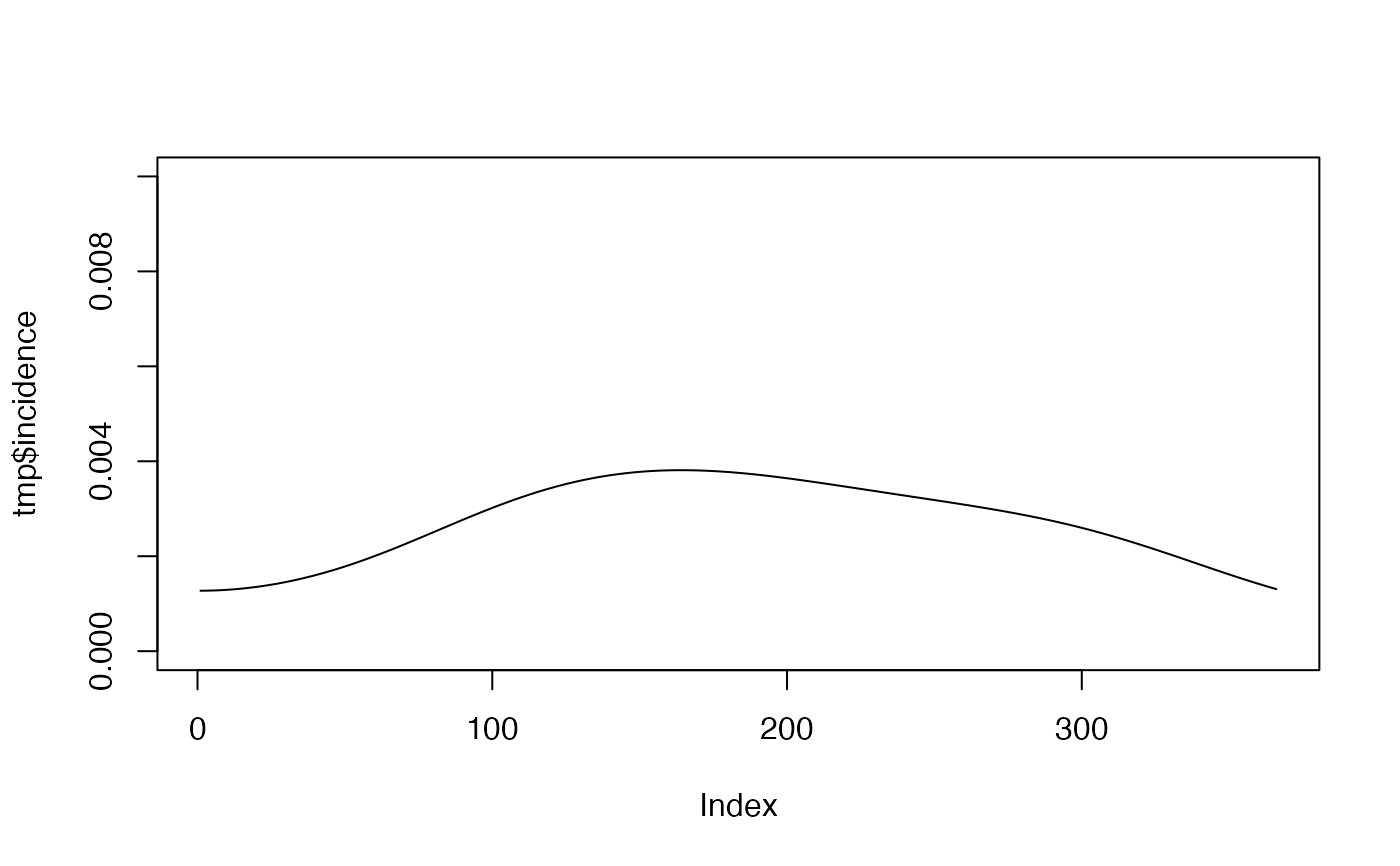
simulate_gaussian_process.RdSimulates an incidence curve (probability of infection per unit time) and associated parameters from a Gaussian Process model assuming that the covariance function (kernel) on time follows the squared exponential.
simulate_gaussian_process(pars)Arguments
- pars
Vector of model parameters, must have entries 1) l: the lengthscale of the covariance function (length of the wiggles); 2) sigma: the output variance (average distance of function from mean); 3) scale_factor: parameter to scale the incidence curve to give a desired "overall probability" of infection (specifically, solves the model, transforms each entry to unit scale, and then scales the entire vector as scale_factor*p/(sum(p))); 4) tmax: the maximum time to solve the model over, generating a time vector starting at 0 and ending at tmax in increments of 1.
Value
a list with two elements: 1) a vector giving the probability of infection at each timestep; 2) a vector giving the parameters used to solve the model